Exploratory Data Analysis Best Practices for Exhaustive Data Analysis
Table of Contents
Introduction
EDA, short for Exploratory Data Analysis, or as I like to call it, Data Reveal Party 😊 (because it’s fun playing around and getting your hands dirty with datasets) is one of the most important steps towards a successful data project. You get to uncover hidden data insights that only this process can unearth. It is therefore good practice to make sure you exhaust on it by investigating and looking at the dataset from every possible angle. In this article we are going to look at the steps to follow to accomplish this when doing EDA.
EDA Steps
1. General dataset statistics
Here you investigate the structure of your dataset before doing any visualizations. General dataset stats may include but are not limited to:
- small overview of dataset - head(), tail() or even sample()
- size and shape of dataset
- number of unique data types
- number of numerical and non-numerical columns
- number of missing values per column
- presence of duplicate values
- column names - correct naming of column names to one word for ease of calling columns from the dataframe object
This is just to familiarise yourself with the dataset. Let’s wrap all these steps in a single python function:
| |
| ID | country_code | region | age | FQ1 | FQ2 | FQ3 | FQ4 | FQ5 | FQ6 | ... | FQ27 | FQ28 | FQ29 | FQ30 | FQ31 | FQ32 | FQ33 | FQ34 | FQ37 | Target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ID_000J8GTZ | 1 | 6 | 35.0 | 2 | NaN | NaN | 2 | NaN | NaN | ... | NaN | NaN | 1.0 | NaN | NaN | NaN | 1.0 | 1.0 | 0 | 0 |
| 1 | ID_000QLXZM | 32 | 7 | 70.0 | 2 | NaN | NaN | 2 | NaN | NaN | ... | NaN | NaN | 2.0 | NaN | NaN | NaN | 1.0 | 2.0 | 0 | 0 |
| 2 | ID_001728I2 | 71 | 7 | 22.0 | 2 | 1.0 | NaN | 2 | NaN | NaN | ... | NaN | NaN | 2.0 | NaN | NaN | NaN | 2.0 | 1.0 | 1 | 0 |
3 rows × 42 columns
shape of dataset: (108446, 42)
size: 4554732
Number of unique data types: 3
ID object
country_code int64
region int64
age float64
FQ1 int64
FQ2 float64
FQ3 float64
FQ4 int64
FQ5 float64
FQ6 float64
FQ7 float64
FQ8 int64
FQ9 int64
FQ10 int64
FQ11 float64
FQ12 int64
FQ13 int64
FQ14 int64
FQ15 int64
FQ16 int64
FQ17 float64
FQ18 int64
FQ19 float64
FQ20 float64
FQ21 float64
FQ22 int64
FQ23 int64
FQ24 float64
FQ35 float64
FQ36 float64
FQ25 int64
FQ26 int64
FQ27 float64
FQ28 float64
FQ29 float64
FQ30 float64
FQ31 float64
FQ32 float64
FQ33 float64
FQ34 float64
FQ37 int64
Target int64
dtype: object
Number of numerical columns: 41
Non-numerical columns: 1
Missing values per column:
ID 0
country_code 0
region 0
age 322
FQ1 0
FQ2 59322
FQ3 62228
FQ4 0
FQ5 87261
FQ6 47787
FQ7 47826
FQ8 0
FQ9 0
FQ10 0
FQ11 24570
FQ12 0
FQ13 0
FQ14 0
FQ15 0
FQ16 0
FQ17 97099
FQ18 0
FQ19 47407
FQ20 24679
FQ21 24635
FQ22 0
FQ23 0
FQ24 70014
FQ35 82557
FQ36 96963
FQ25 0
FQ26 0
FQ27 105246
FQ28 106940
FQ29 24534
FQ30 106331
FQ31 107577
FQ32 47650
FQ33 2
FQ34 31794
FQ37 0
Target 0
dtype: int64
Duplicates: False
Column names: ['ID', 'country_code', 'region', 'age', 'FQ1', 'FQ2', 'FQ3', 'FQ4', 'FQ5', 'FQ6', 'FQ7', 'FQ8', 'FQ9', 'FQ10', 'FQ11', 'FQ12', 'FQ13', 'FQ14', 'FQ15', 'FQ16', 'FQ17', 'FQ18', 'FQ19', 'FQ20', 'FQ21', 'FQ22', 'FQ23', 'FQ24', 'FQ35', 'FQ36', 'FQ25', 'FQ26', 'FQ27', 'FQ28', 'FQ29', 'FQ30', 'FQ31', 'FQ32', 'FQ33', 'FQ34', 'FQ37', 'Target']
After familiarising yourself with the dataset, you can then do some data preparation; data imputation, correcting column names and data types, removing duplicates etc… For our case we see that there are no duplicates but you may find duplicates by subseting columns, say investigating if there are duplicates in the first 10 columns as follows:
| |
| ID | country_code | region | age | FQ1 | FQ2 | FQ3 | FQ4 | FQ5 | FQ6 | ... | FQ27 | FQ28 | FQ29 | FQ30 | FQ31 | FQ32 | FQ33 | FQ34 | FQ37 | Target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3354 | ID_14YEHJ8Y | 4 | 0 | 25.0 | 2 | NaN | NaN | 2 | NaN | NaN | ... | NaN | NaN | 2.0 | NaN | NaN | NaN | 2.0 | 2.0 | 1 | 0 |
| 4133 | ID_1DZ64UE0 | 13 | 0 | 38.0 | 1 | NaN | NaN | 2 | NaN | NaN | ... | NaN | NaN | 2.0 | NaN | NaN | 2.0 | 1.0 | 1.0 | 1 | 0 |
| 4328 | ID_1G9BR3SF | 79 | 0 | 60.0 | 2 | NaN | NaN | 2 | NaN | 2.0 | ... | NaN | NaN | NaN | NaN | NaN | 2.0 | 1.0 | 2.0 | 0 | 0 |
| 5766 | ID_1X42EWSA | 10 | 0 | 30.0 | 1 | NaN | NaN | 2 | NaN | 1.0 | ... | NaN | NaN | NaN | NaN | NaN | NaN | 1.0 | NaN | 1 | 0 |
| 6152 | ID_21OECVBW | 133 | 3 | 55.0 | 2 | 1.0 | NaN | 2 | NaN | 1.0 | ... | NaN | NaN | 1.0 | NaN | NaN | NaN | 1.0 | 2.0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 107939 | ID_ZUKB8ZC6 | 60 | 4 | 37.0 | 2 | NaN | NaN | 2 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | 1.0 | NaN | 1 | 1 |
| 107988 | ID_ZUZ6QUHK | 60 | 0 | 60.0 | 1 | NaN | NaN | 2 | NaN | NaN | ... | NaN | NaN | 2.0 | NaN | NaN | NaN | 2.0 | 1.0 | 1 | 0 |
| 107994 | ID_ZV2EK93X | 45 | 4 | 49.0 | 2 | NaN | NaN | 2 | NaN | 1.0 | ... | NaN | NaN | 2.0 | NaN | NaN | 2.0 | 1.0 | 1.0 | 1 | 0 |
| 108313 | ID_ZYKSG3XG | 131 | 1 | 46.0 | 1 | 1.0 | NaN | 2 | NaN | NaN | ... | NaN | NaN | 2.0 | NaN | NaN | NaN | 1.0 | 1.0 | 0 | 0 |
| 108379 | ID_ZZ7M49OG | 45 | 3 | 53.0 | 1 | 1.0 | NaN | 2 | NaN | 1.0 | ... | NaN | NaN | 2.0 | NaN | NaN | 2.0 | 1.0 | 1.0 | 1 | 1 |
645 rows × 42 columns
That way we find that there are 645 duplicate rows in the dataset. You can use this convention to flag columns that say, shouldn’t have any repetitions as a requirement.
2. Univariate Analysis
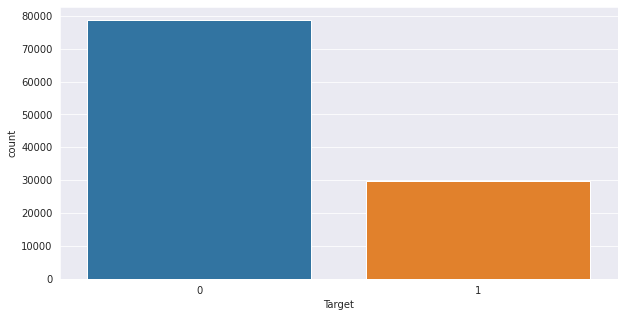
Univariate; analysis of one variable. Here we investigate trends per single column of interest. Keyword here is uni/one/single. We take a column of interest and investigate patterns in it before going to the next. For example we may want to know how the target variable in our sample dataset above is distributed, or the youngest and oldest customer and age group thresholds(definite age groups as seen in histogram bars). We do analysis column by column.
| |
0 78735
1 29711
Name: Target, dtype: int64
<AxesSubplot:xlabel='Target', ylabel='count'>
| |
(array([30748., 30169., 22129., 17240., 6919., 919.]),
array([15., 29., 43., 57., 71., 85., 99.]),
<BarContainer object of 6 artists>)
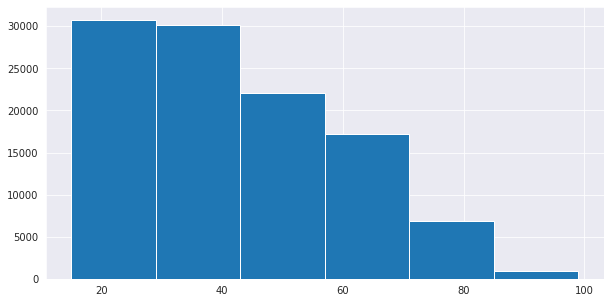
This information from the age column can be used to create a categorical variable as a feature engineering practice.
3. Bivariate Analysis
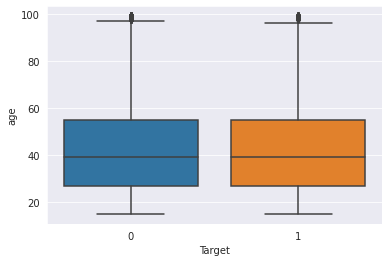
You guessed it! Here we do analysis of two variables. We try to determine the relationship between them. One type of plot that I never miss to use under bivariate analysis is the boxplot, especially in helping one to detect and remove outliers. It also helps one to visually see quartiles and their ranges.
| |
<AxesSubplot:xlabel='Target', ylabel='age'>
| |
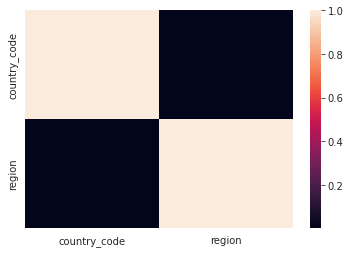
country_code region
country_code 1.000000 0.002045
region 0.002045 1.000000
<AxesSubplot:>
| |
<seaborn.axisgrid.JointGrid at 0x7f48697e3400>

| |
<matplotlib.collections.PathCollection at 0x7f48684f0370>
4. Multivariate Analysis
This involves observation and analysis of more than two variables at a time. We can also add onto bivariate analysis observations at this step by including the hue parameter to take care of a third distingushing feature between the two.
| |
<matplotlib.collections.PathCollection at 0x7f48681d6520>
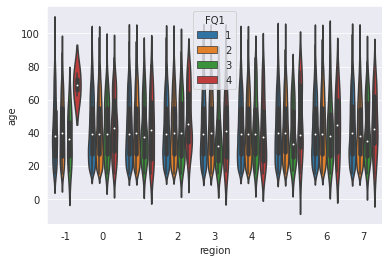
| |
<AxesSubplot:xlabel='region', ylabel='age'>
To finish off multivariate analysis, there is one, many in one, plot that carries a lot of information from all the variables; the pairplot. For ease of readibility we’ll include only the first few columns together with the target variable since our dataset has a lot of variables:
| |
<seaborn.axisgrid.PairGrid at 0x7f4861c7c160>

Most of the plots inside the pairplot don’t tell a lot of information in our case because these many variables are categorical. You should try it with a more receptive dataset.
These are the steps for exhaustive data analysis. We went through each giving a few examples. This guide does not cover everything, but it outlays a good procedure to follow for exhaustive EDA. It is good practice to go into details and make as many beatiful visualizations at each step, cheers!